Hydrogen-deuterium exchange mass spectrometry is a mass spectrometry technique that studies the spatial conformation of proteins. It has a wide range of applications in the study of protein structure and dynamic changes, discovery of protein interaction sites, identification of protein epitopes and active sites. With the continuous development of hydrogen deuterium exchange mass spectrometry, it is becoming an important means for structural biologists and biopharmaceutical research and development.
Hydrogen deuterium exchange mass spectrometry (HDX MS) is a mass spectrometry technique that studies the spatial conformation of proteins. The principle is that the protein is immersed in the heavy aqueous solution. The hydrogen atoms of the protein will exchange with the deuterium atoms of the heavy water, and the hydrogen in close contact with the heavy water on the protein surface is faster than the hydrogen located inside the protein or participating in the formation of hydrogen bonds Mass spectrometry was used to determine the hydrogen-deuterium exchange rate of different protein sequence fragments, thus obtaining protein spatial structure information [1]. This process is like immersing a fist in the water, and then raising the surface of the water and opening the palm. At this time, the back of the wet hand indicates that it is on the outer surface of the "fist" structure, while the drier palm indicates that it is inside the "fist". In addition to sample preparation, the main processes of hydrogen deuterium exchange mass spectrometry include: exchange reaction, termination reaction, rapid protein digestion into peptides, liquid phase separation, mass spectrometry detection, data analysis. Among them, the exchange step needs to be performed under multiple reaction time, such as 0s, 10s, 1min, 10min, 60min, etc., in order to draw the exchange rate curve to obtain accurate and comprehensive information. Hydrogen-deuterium exchange mass spectrometry has a wide range of applications in the study of protein structure and its dynamic changes [1], discovery of protein interaction sites [2], identification of protein epitopes and active sites [3].
Compared with classical protein structure research methods, such as X-Ray Crystallography and Nuclear Magnetic Resonance (NMR), hydrogen deuterium exchange mass spectrometry cannot provide accurate protein spatial structure. It directly provides The main information includes which amino acid sequences are located on the surface position of the protein spatial structure (including dynamic changes), possible active sites and protein-protein interaction sites. However, hydrogen deuterium exchange mass spectrometry has advantages that other classic methods do not have: first, the study of dynamic changes in protein structure is an outstanding advantage of hydrogen deuterium exchange mass spectrometry, including the changing active sites and epitopes; second, hydrogen deuterium Exchange mass spectrometry also has unique advantages in the study of protein complex conformation; in addition, hydrogen deuterium exchange mass spectrometry also has a small sample demand, relatively low purity requirements, and the research object is the natural conformation of the protein in a solution environment instead of crystal Advantages such as medium conformation [1,4,5]. Since the first research paper was published in 1991, hydrogen deuterium exchange mass spectrometry technology has been continuously developed and has become a very important application field in structural biology and mass spectrometry technology [6]. However, the complex realization process of the hydrogen deuterium exchange mass spectrometry experiment has affected its application to a certain extent. The main difficulties are: 1. How to avoid the backcross phenomenon of deuterated peptides after the exchange; 2. The high accuracy and reproducibility requirements of the experimental control; 3. How to accurately distinguish the superimposed mass spectrum peaks caused by the exchange; 4. Simple and efficient analysis software requirements; 5. Identification of exchange sites in amino acid units. Since 2005, Waters has continued to tackle the above difficulties and launched the only commercial fully automated hydrogen deuterium exchange mass spectrometry system solution-nanoACQUITY UPLC? HD-Exchange System (Figure 1). Worldwide, this system has helped scientists publish research papers in top research journals including Cell and Nature [7,8]. In addition to scientific research needs, Waters hydrogen deuterium exchange mass spectrometry system has also been recognized by many leading international pharmaceutical companies and is used in the research work of protein drug active sites and epitopes in new drug development.
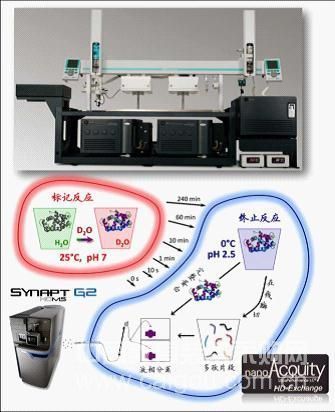
Figure 1. nanoACQITY UPLC HD-Exchange System.
The backcross phenomenon in the hydrogen-deuterium exchange experiment will seriously affect the credibility of the experimental data, and even lead to erroneous results. To avoid backcrossing, two points need to be achieved: try to shorten the liquid quality analysis time and ensure that the temperature and pH in the liquid quality analysis are the environments required by the minimum backcross reaction coefficient. Waters UPLC? System uses sub-two nano-chromatographic particle packing. Compared with the large particle packing used by HPLC, UPLC has unparalleled resolution. Therefore, UPLC can achieve the requirement of greatly shortening the liquid phase analysis time without losing the chromatographic separation effect [9]. For the control of temperature and pH, in the years of engineering improvement, nanoACQUITY UPLC HD-Exchange System has achieved full control of enzyme digestion and liquid phase separation [10].
The requirement of the accuracy and reproducibility of the hydrogen deuterium exchange mass spectrometry experiment is the second major difficulty of its application. In the experiment, it is generally necessary to collect data at multiple time points such as 0s, 10s, 1min, 10min, 60min, and 240min. If manual manual experiments are performed, it is difficult to perform precise operations at several time points such as 10S-10min. Taking into account the need to repeat experiments, manual manual operations will have an impact on the credibility of the final data. Moreover, the repetitive and cumbersome experimental process will bring great pressure to the experimental staff. The nanoACQUITYUPLC HD-Exchange System completely completes a series of experimental processes such as exchange, termination of exchange, injection, and enzyme digestion through an intelligent robotic arm, and always guarantees different temperature environments required for each step. These automated processes not only ensure the reliability of the experimental data, improve the efficiency of the experiment, but also liberate the scientists from the tedious and repeated experiments.
In the mass spectrometry data of the hydrogen deuterium exchange experiment, with the extension of the exchange time, the peptide that has undergone the exchange reaction will gradually move toward the direction of high mass-to-charge ratio due to the increase in mass. Therefore, these mass spectrometry peaks may gradually overlap and overlap with the peptide mass spectrum peaks that have not undergone exchange reactions. The mass spectrum signals superimposed on each other not only affect the judgment of peak assignment, but also increase the error of the exchange rate data. Because the exchange rate judgment needs to be quantified by the peptides that are exchanged, there is no doubt that the superimposed and chaotic mass spectrometry data will greatly affect the accurate quantification of the mass spectrum peaks. This is completely useless for mass spectrometers that analyze purely by mass-to-charge ratio. However, this seemingly impossible task was overcome by the Waters nanoACQUITY UPLC HD-Exchange System. This is because, unlike other common mass spectrometers, Waters' SYNAPT? Mass spectrometry platform also has the function of separation based on ion size and morphology (travel wave ion mobility separation). In data processing, in addition to the mass-to-charge ratio information of peptide ions, different ions can also be distinguished by ion migration time (ion mobility dimension parameter). Therefore, this unique SYNPAT mass spectrometry technique named HDMSE can separate peptides that overlap due to the same mass-to-charge ratio, easily solve the problem of superposition of mass spectrometry signals, and obtain accurate exchange rate data [11,12] (figure 2). Once launched, the SYNPAT mass spectrometry platform won the 2007 PITTCON Gold Award. At present, it has launched a new generation of SYNAPT G2HDMS, SYNAPT G2-S HDMS and other models, and has multiple ion sources such as ESI and MALDI. In addition to the hydrogen-deuterium exchange technology, the SYNAPT mass spectrometry system is also unique in the study of protein complex structures, and many high-quality applications have been published [13,14,15].

Figure 2. The top four images in the figure are the mass spectra of the polypeptide YTNHTVLPEAL2 + and the polypeptide IITAGDVVNHDPVVGDRL3 + under different hydrogen-deuterium exchange reaction time. It can be seen from this figure that as the exchange time increases, the mass spectrum signals of the two peptides gradually overlap together. The following two images are obtained by using the HDMSE mass spectrometry acquisition method. The mass spectrum peaks covered by the two peptides are separated by different ion mobility migration time.
[Liquid quality analysis method of protein and peptide]
The fourth key point to realize hydrogen deuterium exchange mass spectrometry technology is how to efficiently analyze the large amount of data brought by multiple time points and multiple repetitions generated by the experiment. It takes a lot of time for scientists to complete such huge information processing work manually. The DynamX software provided by Waters hydrogen deuterium exchange mass spectrometry solution can provide scientists with simple and intuitive analysis results, and includes multiple presentation methods.
In some special studies, the measurement of protein hydrogen deuterium exchange sites is required to be accurate to the measurement of amino acids, which is another difficulty in the study of hydrogen deuterium exchange mass spectrometry. The use of CID (Collision Induced Dissociation) fragmentation mode in routine research may lead to rearrangement of deuterium atoms in the polypeptide, which makes it impossible to pinpoint the specific amino acids that are exchanged. The ETD (electron transfer dissociation) fragmentation mode provided by SYNPAT mass spectrometry can avoid the information confusion caused by the rearrangement of deuterium atoms, and has a good fragmentation signal [16].
Waters' nanoACQUITY UPLC HD-Exchange System provides an unprecedented and simple solution for hydrogen deuterium exchange mass spectrometry experiments, which strongly promotes the hydrogen deuterium exchange technology in protein structure and dynamic change research, protein interaction site discovery, protein The application of epitope and active site identification is becoming an indispensable work platform for many structural biology scientists and biopharmaceutical companies.
references
(1) John R. Engen, Analysis of Protein Conformation and Dynamics by Hydrogen / Deuterium Exchange MS. Anal. Chem. 2009,81, 7870–7875
(2) Engen et al. Probing protein interactions using HD exchangems in ms of protein interactions. Edited by Downard, John Wiley & Sons, Inc. 2007, 45-61
(3) Tiyanont K, Wales TE, Aste-Amezaga M, et al. Evidence forincreased exposure of the Notch1 metal oprotease cleavage site uponconversion to an activated conformation. Structure. 2011, 19, 546-554
(4) Heck AJ. Native mass spectrometry: a bridge betweeninteractomics and structural biology. Nat Methods. 2008, 5, 927-933.
(5) Esther van Duijn, Albert JR Heck. Mass spectrometricanalysis of intact macromolecular chaperone complexes. Drug Discovery Today. Drug Discovery Today: Technologies Volume 3, 2006, 21-27
(6) Viswanat ham Katta, Brian T. C hait, Steven Carr.Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1991, 5, 214–217
(7) Chakraborty K, Chatila M, Sinha J, et al. Chaperonin-catalyzed rescue of kinetical y trapped states in protein folding. Cel. 2010 Jul9; 142 (1): 112-22.
(8) Zhang J, Adri 醤 FJ, Jahnke W, et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010,463, 501-50636
(9) Wu Y, Engen JR, Hobbins WB. Ultra performance liquid chromatography (UPLC) further improves hydrogen / deuterium exchange mass spectrometry. J Am Soc Mass Spectrom. 2006, 17, 163-167
(10) Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008, 80, 6815-6820
(11) Giles K, Pringle SD, Worthington KR, et al. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004, 18, 2401-2414
(12) Olivova P, C hen W, C ha kra borty AB, Gebler JC. Determination of N-glycosylation sites and site heterogeneity in a monoclonal antibody by electrospray quadrupole ion-mobility time-of-? Ight mass spectrometry. Rapid Commun Mass Spectrom. 2008, 22,29-40
(13) Ruotolo BT, Benesch JL, Sandercock AM, et al. Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc. 2008, 3, 1139-52.
(14) Uetrecht C, Barbu IM, Shoemaker GK, et al. Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat Chem. 2011, 3,126-132
(15) Bleiholder C, Dupuis NF, Wyttenbach T, Bowers MT. Ion mobility-mass spectrometry reveals a conformational conversion from random assembly to β-sheet in amyloid? Bril formation. Nat Chem. 2011, 3, 172-177
(16) Kasper D. Rand, Steven D. Pringle, Michael Morris, John R., et al. ETD in a Traveling Wave Ion Guide at Tuned Z-Spray Ion SourceConditions Allows for Site-Specific Hydrogen / Deuterium Exchange Measurements. J Am Soc Mass Spectrom. 2011, in press
Stop the finishing rolls coming from the rewinder, then free them to the V-slat conveyor in sequence and interval. Let paper rolls be conveyed I the preset order.
Up till now, we have developed many new systems like Palletizer, strapper, paper corner applicator and Carton Sealer, etc. We are already ready to serve you with our best.
Stop Sorting Deck,Paper Sorting Machine,Stopping Sorting Deck For Paper Roll
Shandong Sinolion Machinery Corp., Ltd. , https://www.sinolion.cc